Image-Based Cell Profiling and Data Analysis
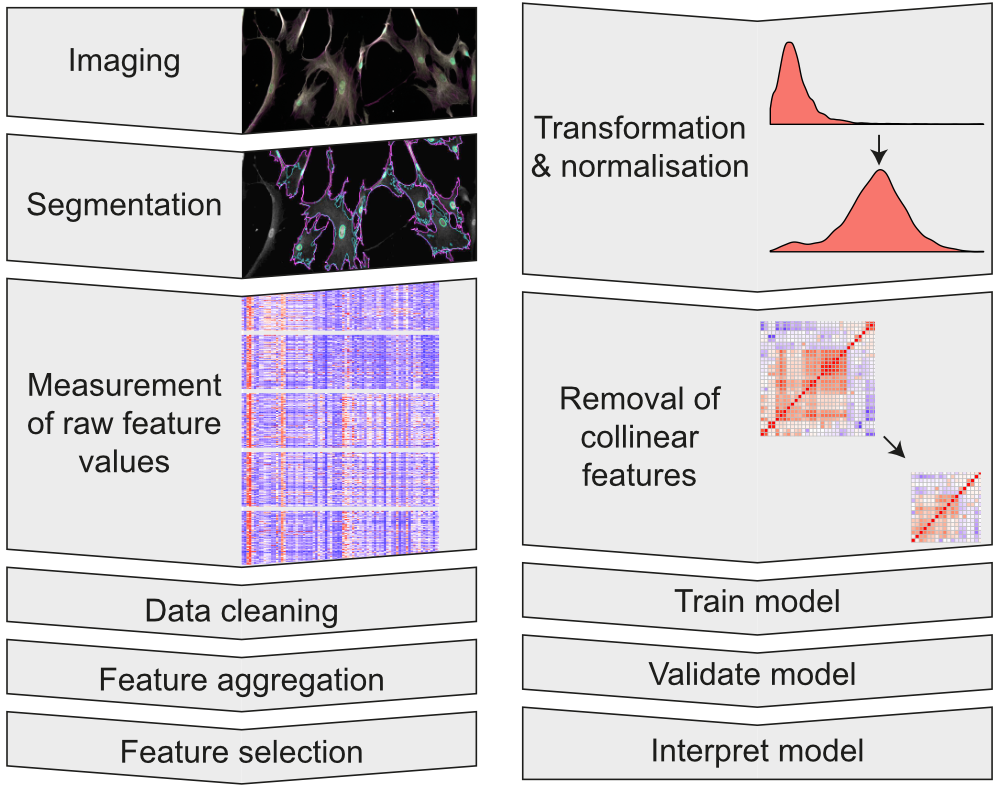
We work with large microscopy datasets and image-based cell profiling tools to automate the process of analysing hundreds of thousands of cells.
Our methods
We primarily use Ilastik and CellProfiler to perform pixel classification and image segmentation, to generate our initial datasets. We then clean, analyse and model our data using the R programming language in order to generate insight and meaningful outcomes.
Quantifying the response of cells to high-aspect-ratio nanostructures
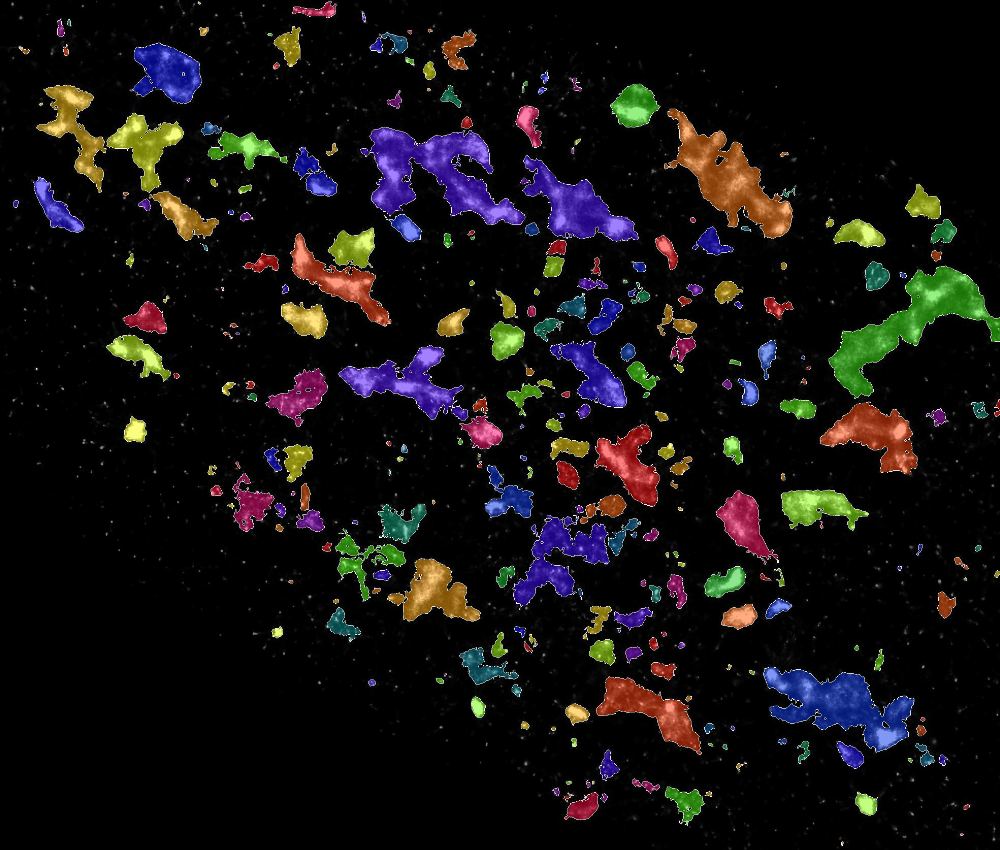
To uncover the complexity of cellular response to high-aspect-ratio nanostructures, we construct image analysis pipelines. These can process single-cell measurements from hundreds of thousands of cells from each experiment.
Assessing many cells at once allows us to understand how the distributions of morphological features and staining intensities vary across a heterogeneous cell population, revealing subtle effects.1
Download Stuart's workshop on working with CellProfiler data in R
Analysing the response of progenitor cells in a neural tube model
Working as part of the PhD project of Dr Daniel Boland, and in collaboration with Dr Despina Stamataki of the Briscoe Lab, The Francis Crick Institute, Stuart constructed an image analysis pipeline to assess the state of neural mesodermal progenitor cells in a neural tube model.
Stuart developed pipelines to assess the ventral/dorsal state of the progenitor cells. This approach allows the separation of background and density effects from the model system.
Analysing the geometry of lysosomes from STORM images
Stuart worked to analyse the impact of a new therapeutic on the geometry of lysosomes, supporting the work of Dr Jeremy Bost and Dr Miina Ojansivu at the Karolinska Institutet, Sweden. Stochastic Optical Reconstruction Microscopy (STORM) images were analysed using a combination of Ilastik / CellProfiler pipleline, to overcome the limitations of thresholding based pixel classification. The analysis revealed subtle effects of the therapeutic on lysosome geometry.2
-
Adapted under terms of CC-BY license from: Seong H, Higgins SG, Penders J, Armstrong JPK, Crowder SW, Moore AC, Sero JE, Becce M, Stevens MM, Size-Tunable Nanoneedle Arrays for Influencing Stem Cell Morphology, Gene Expression, and Nuclear Membrane Curvature, 2020, ACS Nano. DOI: 10.1021/acsnano.9b08689 ↩
-
Adapted under terms of CC-BY license from: Bost JP, Ojansivu M, Munson MJ, Wesén E, Gallud A, Gupta D, Gustafsson O, Saher O, Rädler J, Higgins SG, Lehto T, Holme MN, Dahlén A, Engkvist O, Strömstedt P-E, Andersson S, Smith CIE, Stevens MM, Esbjörner EK, Collén A, El Andaloussi S, Novel Endosomolytic Compounds Enable Highly Potent Delivery of Antisense Oligonucleotides, 2022, Commun. Biol. DOI: 10.1038/s42003-022-03132-2 ↩